CDCB Genomic Dictionary¶
Blend code¶
It describes whether the reference population used for evaluation was composed of a single or multiple breeds.
| Blend Code | Description |
| S | Single breed/no blending |
| M | Multiple breed/blended |
| X | Non-genomic breed or breed conflict |
| P | Pending. Genotype not yet processed for BBR |
Breed base representation (BBR)¶
Breed base representation (BBR) was introduced in 2016. Most crossbreds have not been included in genomic evaluations because marker effects are computed separately within breeds. Edits that determine which animals are evaluated use a small set of breed-check markers (see Breed conflict determination below). Using all markers allows each animal's ancestry to be estimated more precisely. Breed base representation estimates the percentage of DNA contributed to the animal by each of 5 evaluated breeds: Holstein, Jersey, Brown Swiss, Ayrshire, and Guernsey. These 5 new fields sum to 100 (with a minimum of 0 and a maximum of 100). BBR values of 94 to 99% are set to 100% because such values occur often even for animals with 100% purebred ancestry. The initial BBR estimates have a standard error of about 2% caused by normal variation within a breed as well as additional error caused by imputation from lower density chips. BBR values are distributed once a month for each animal.
To calculate BBR we solve for individual SNP effects the same as we do to get direct genomic values for any trait. In this case, the Y = 100 for members of the purebred reference population being estimated and 0 for the other breeds. A set of SNP effects is estimated for each of the 5 breeds with appropriate changes in the value of the Y's. Then for each animal the Direct Genomic Value (DGV) is calculated for each breed.
BBR reference Population¶
The BBR reference population is selected from the genotyped, progeny-tested bull population who are enrolled (excluding bulls with status codes C and N) with complete pedigree and classified as purebred within each breed of evaluation. Purebred bulls must have a pedigree-based heterosis value ≤ 1% (rounded to the nearest integer). A complete pedigree is defined as a pedigree with all ancestors identified for four generations. Animals with constructed IDs in their four-generation pedigree are disqualified.
Exceptions:
- All breeds (except Jersey): Bulls born before 2000 with more than 1,000 daughters in Holstein or more than 200 daughters in other breeds are exempt from the pedigree completeness rule. This accommodates older but influential bulls with partially complete pedigrees. Per an agreement with AJCA, this rule does not apply to the Jersey breed.
- Jersey breed: All animals with “JX” at the beginning of their name are automatically disqualified from the reference population.
- Brown Swiss breed: Intergenomics bulls with MACE proofs are treated equivalently to enrolled progeny-tested bulls.
Benefits of BBR¶
- BBR values represent the breed composition of an individual that is more accurate and much easier to interpret than breed-check markers
- BBR provides a method for combining the marker effects from different breeds into accurate genomic predicted transmitting abilities (GPTAs) for crossbreds.
- The use of BBR provides the means for making genetic predictions for crossbreds possible.
Publications and other resources:¶
- VanRaden, P.M., and T.A. Cooper. 2015. Genomic evaluations and breed composition for crossbred U.S. dairy cattle. Interbull Bull. 49:19–23. | PowerPoint presentation
- Norman, H.D., VanRaden, P.M., Megonigal, J.H., Durr, J.W., and Cooper, T.A. Breed base representation in dairy animals of five breeds. J. Dairy Sci. 99(E–Suppl. 1)/J. Anim. Sci. 94(E–Suppl. 5):151–152(abstr. 0324). 2016.
- VanRaden, P.M., Olson, K.M., Wiggans, G.R., Cole, J.B., and Tooker, M.E. Genomic inbreeding and relationships among Holsteins, Jerseys, and Brown Swiss . J. Dairy Sci. 94(11):5673–5680. 2011.
- https://www.usjersey.com/Portals/0/AJCA/2_Docs/Animal-Applications/Norman-BBR-Seminar.pdf
- https://hoards.com/article-18985-usjersey-today-bbr-released-with-monthly-genomic-evaluations.html
- https://www.icar.org/Documents/Auckland-2018/1430%20Dr%20Duane%20Norman%20CAR%20New%20Zealand.pdf
Breed conflict determination¶
Breed conflict determination is performed at the time of processing of the gentoype. There are several criteria used in determining a breed conflict
- Breed of ID is not HO/JE/AY/GU/BS/XX (see "Note1")
- A different eval breed has < 10% unlikely breed SNP alleles.
- No evaluation if the max(BBR) > 89.5 and not for the eval breed.
Note1: AY/SR/NR/RE are processed as AY
Note2: Breed SNP are also used to determine which genotypes should be processed in the XX directory which is indicated by conflict code "l" which is assigned when the unlikely allele is present for > 15% of the breed SNP for the eval breed.
Clone genotype¶
An animal with identical nuclear DNA as the animal with the DNA ID (defined as the animal with common nuclear DNA of the clonal family) through embryo splitting or nuclear transfer. Natural identical twins also can be accommodated by this system if their common DNA is confirmed.
Constructed dam IDs¶
IDs with DAM or MGD following the country code (ex. HOUSADAM087654321) - When a likely grandsire or great grandsire can be suggested based on the percentage of haplotypes in common with the genotyped animal, and the dam or granddam is unknown, an ID is constructed to link the discovered male ancestor to the genotyped animal.
Crossbred Evaluation¶
The CDCB calculates genomic predictions on over 40 different traits separately for 5 different dairy breeds: Ayrshire, Brown Swiss, Guernsey, Holstein, and Jersey. Genomic predictions for animals that have ancestors of more than one of these 5 breeds are calculated as a blended average of the respective single-breed marker effects, weighted by the estimated portion of the animal's DNA that came from each of the 5 breeds. (detailed info available at Genomic evaluations including crossbred animals policy)
The evaluation system was designed based on complex rules, policies, and conditions as stated in the documentation above.
The flow of determining the breed of evaluation and obtaining genomic evaluations including crossbred animals are described below: 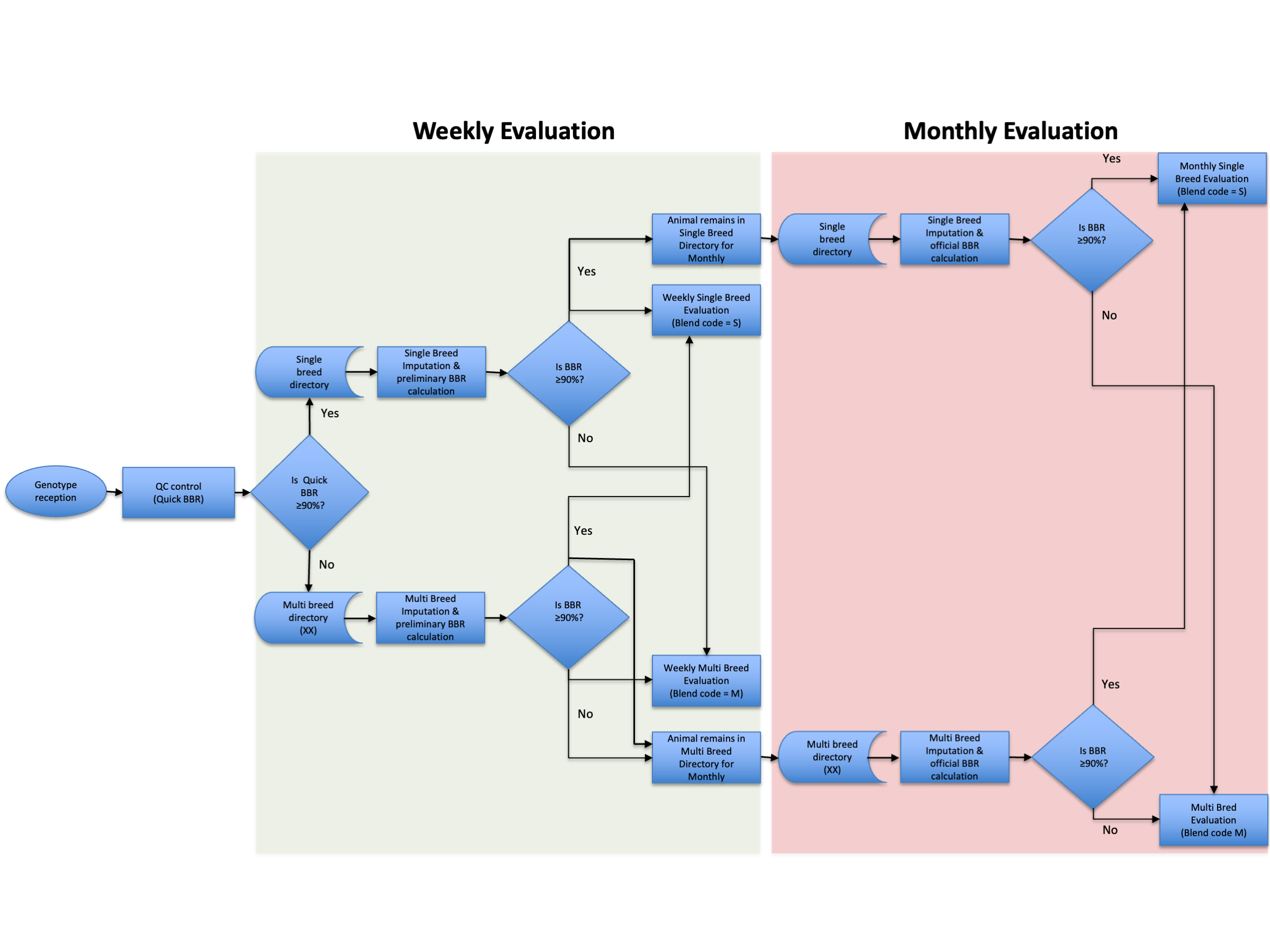
Dairy Record Processing Center (DRPC)¶
Data at the farm level is typically collected by a Dairy Herd Improvement (DHI) center. The data is then submitted to a Dairy Record Processing Center (DRPC) for normalization and secondary processing. The DRPC then send the applicable records to CDCB, which performs a high-level editing of the recods received. The data submitted by DRPC to CDCB are: lactation, reproduction, health, calving, herd, test day, yearly average, etc.
When pedigree records have source code "D", it means it the record was submitted by a DRPC's. In this case, DRPCs are responsible to correct the records, and should be contacted directly:
| Center No | Location | Contact person | email address |
|---|---|---|---|
| 01 | CA | Lisa Pierucci Teresa Parreira |
lpierucci@agritech.com tparreira@agritech.com |
| 07 | NC | Tammie Guyer Greg Palas |
tammie_guyer@ncsu.edu gepalas@iastate.edu |
| 10 | UT | Michelle Gren | michelleg@amelicor.com |
| 12 | WI | Customer Service (Record correction) Rachel Hellenbrand Melinda Pegram |
custserv@agsource.com rachel.hellenbrand@agsource.com melinda.pegram@agsource.com |
Embryo genotypes¶
CDCB accepts genotypes from embryos with the same QC sample quality thresholds as for live animals.In terms of the nomination procedure, there are 2 main differences:
- CDCB requires the nominator to use " EMBRYO " as the name of the animal at @100-129 in Format1 or in the Nomination Query (requires login).
- CDCB requires the nominator to use code '6' in the multiple birth code at @91 in Format1 or in the Nomination Query (requires login). This requirement is important as it prevents the "birthdate" from conflicting with the dam birth date.
- Failure to report embryo genotype correctly will result in the embryo information not being merged to the live animal when the live animal is loaded into the CDCB database
- Embryos are routinely withdrawn after notifying the nominator when matched with a live calf, or when 2 years have passed since they were stored in our database.
- If you do not want the withdrawal to occur, change the multi-birth code if it is actually a live calf, or change the name to Cell line to prevent the withdrawal.
Cell line genotypes¶
CDCB also accepts genotypes from cell lines, similar to embryo genotypes.In the nomination,
- CDCB requires the name field (@100-129 ) in format1 to consist of Cell line. This input is case sensitive. The name cannot contain anything other than Cell line.
- CDCB WILL NOT withdraw the cell-line genotypes after 2 years, unlike embryo genotypes.
Genomic Inbreeding (Gen_Inb)¶
Gen_Inb measures the actual homozygosity and percentages of genes in common of an animal. The genomic inbreeding is obtained from the diagonal of the genomic relationship matrix and is related to the portion of an animal's markers that are homozygous. Pedigree inbreeding or relationship of parents does not measure the Mendelian Sampling component of which haplotypes are inherited, and thus all full sibs have the same pedigree relationship and inbreeding, whereas genomic inbreeding is much more precise by determining which markers in an animal are homozygous.
Mendelian inheritance conflicts¶
Mendel's law of segregation states that, during gamete formation, alleles segregate so that each gamete carries only one allele for each gene. Consequently, the progeny of two individuals (e.g. the sire and the dam) is expected to carry one copy of alleles from each parent.
This means that an individual with an homozygous SNP (e.g. AA) will create two gametes carrying the same allele (A), which means his progeny will receive at least one allele A for that SNP. If the progeny of the individual is, however, homozygous for the alternative allele (e.g. BB) this is considered a conflict to the aforementioned law.
A few examples:
| PARENT genotype | PROGENY genotype | CONFLICT |
|---|---|---|
| AA | AA | NO |
| AB | AA | NO |
| BB | AA | YES |
| AA | AB | NO |
| AA | BB | YES |
The main causes for these conflicts are:
- A) genotyping error. The SNP in the parent or the progeny was actually heterozygous (e.g. AB) but wrongly assigned as homozygous. The SNP chip genotyping technology is quite accurate and the amount of such errors is quite low, therefore only a handful of these errors are expected (and tolerated).
- B) wrong pedigree. A large number of such conflicts is evidence that the parent-progeny relationship is not correct.
Negative Key¶
Negative Key is assigned to a genotype that is conflicting with another genotype that is assigned to the same animal. A negative key gets assigned automatically by our system especially when the genotype is determined as less likely to be the animal's. For example, if one genotype has confirmed parents and another one has parent-progeny conflict, then one with the conflict will be assigned the negative key. The solution to the negative key is to find the correct animal and move the negative key genotype the correct animal.
Notify file/Format1E¶
After a format1 record is submitted to the “in” directory, the nominator should check whether there is a format1E file (YYYYMMDD.1EX) or a notify file (notify.20170418.1X) placed in the “out” directory. These files will contain indications of the nomination process (or indicate the type of corrections required).
In case of Format1 submission, these error files are created only in case of presence of error codes, whereas for Format 1G submission these files are always created as they contain a record for each nomination (successful/unsuccessful nomination report).
Format1E file (YYYYMMDD.1EX)
This file is very similar to format1 except that there are some error information after byte position 141 (Error code information, conflicting or changed identification, and herd code information). A detailed description of this format is available at Format 1E format information.
The most important information in this file is the error code(s) that provides indications on errors in the submission. Error codes (consists of one number, one capital letter and one small letter like “1Ab”) are available at byte positions: 142-144, 179-215, 216-252, 253-289, 290-326, and 327-362, depending on how many errors the submisison had.Error code documentation is available at https://queries.uscdcb.com/formats/geterr.cfm
Notify file (notify.YYYYMMDD.1X)
This file contains exactly the same information of a Format 1E file, but in human-readable format.
Errors and notifications provided by CDCB system from the processing of Format1 submissions are shown in this file only when there are errors. Among the notifications, status of the nominations resulting from Format1G processing are always shown in this file ("Nomination Acceptance" field). An example is attached (Notify_ex.JPG) at the bottom of this page.
Specifically, the notification area has 7 columns that provide all the information resulting from the processing of a record:
- "Code": this is a 3-letter code of the information provided. Detailed description is available at https://queries.uscdcb.com/formats/geterr.cfm as well as brief description is indicated under "Reason" field in the notify file
- "Dsp": is a 1-letter code, that indicates the disposition Code (e.g. type of information provided).
- "Reason": brief description of the code. If the description is not sufficient, more detailed information is available at the Error documentation .
- "ID": an ID (such as dam, sire, etc) involved in the error, if applicable. This information is provided to help determining the cause of the error (eg, calving date of the dam does not match with the birth date of the animal, therefore the dam's ID is indicated in this field).
- "Date": date of the event involved in the error, if applicable. This information is provided to help determining the cause of the error (eg, calving date of the dam does not match with the birth date of the animal, therefore the calving date of the dam is provided in this field).
- "Herd": Herd of the animal that is involved in the error is indicate, if applicable. This information is provided to help determining the cause of the error (eg, calving date of the dam does not match with the birth date of the animal, therefore the herd ID of the dam is provided in this field).
- "Source": source code of the record involved in the error segment. However, this field also returns "Y" or "N" as the as a result of the "Nomination acceptance" field (nomination accepted or not). As an example, "Y" for accepted or N" not accepted can be indicated when format1G was submitted for nomination. The attached example (Notify_ex.JPG) contains "N" along with error code "0Ri" in the source field. This means that the nomination failed for that record.
What notify file contains the information needed when format1G is submitted?
When format1G is submitted, the CDCB system performs a parentage verification and a nomination confirmation.
In the notify file, if the parentage verification succeeded, only the nomination confirmation information is included. However, if the pedigree was not verified or needs your attention, then the notify file will also include the error information from the parentage verification.
There are two files notify files named “notify_VAL.YYYYMMDD.1[G]E” and “notify_ERR.YYYYMMDD.1[G]E”, respectively. Note that since format 1’s can be used for multiple purposes (and multiple records for a single animals could be sent), the same animal could be present in both files. The “Format 1[G]E” file will still be delivered to provide a more “script-able” format to our collaborators.
notify_VAL.YYYYMMDD.1GX
Notify whether the animal was nominated successfully or not regardless of existence of errors.
notify_ERR.YYYYMMDD.1[G]X
Notify only if error/change needs to be informed.
Sex abnormality¶
Since males have one X and one Y chromosome and females have two X chromosomes, the system expects no heterozygous calls in the X chromosome in males and some calls on the Y chromosome. Similarly, females are expected to have some heterozygous calls in X chromosomes and no calls on the Y chromosome. If an animal has a higher than expected number of calls for its declared sex, the CDCB system will report a sex conflict. Sex conflicts prevent those animals to be evaluated. However, in exceptional cases and if valid reasons or evidences that support the results obtained, CDCB can include animals into our evaluation . Since only CDCB staff can implement those exceptions, the nominator needs to contact the CDCB customer service using a Redmine ticket.
| Sex | X Heterozygosity | Y-Count | Valid Y-Count, Invalid Hetero |
| For Males: | hetero <1.4 is valid, 1.4-7.2% is abnormal, >7.2% is a sex conflict | Must have >70% available Y-SNPs | The genotype is considered unreliable, possible XXY |
| For Females: | hetero <7.2% is sex conflict†, 7.2-17.2% is abnormal, >17.2% is valid | Must have <20% available Y-SNPs | Considered valid, but unusual‡ |
†If chip has no Y-SNPs and female has common ancestors, the sex is considered valid, but unusual when X hetero is <7.2%
‡If the female does not have valid Y-count (ex. 3-4), the genotype is considered unreliable
Unlikely and Discovered Grandsires¶
Single-SNP analysis¶
The Single-SNP analysis is used for grandsire (GS) search when the sire is not confirmed. It is also used to determine if a GS is unlikely.
Effective in 2017, animals with unlikely GS are excluded from the evaluations. The percentage of SNP conflicts required for the GS to be unlikely ranges from 29% to 18%. The unlikely GS conditions are distributed html format after each genotype submission as well as displayed in the Genotype Query.
Threshold calculation
The actual threshold depends on the number of SNP that were checked. The SNPs will be checked if either the animal and the MGS are homozygous for the SNP, or both the sire and MGS are genotyped, and both are homozygous for the same allele. The regression equation is
threshold = 32.70071 -0.00184*checked; giving
3000 27.2
4000 25.3
5000 23.5
6000 21.7
7000 19.8
8000 18.0
The limits, max 29% for < 2000 checked and min 18% for > 8000 checked are imposed.
Timing: Single-SNP analysis is used whenever the genotype is processed to suggest possible MGS for genotypes without confirmed sire and unlikely MGS
Haplotype Based analysis¶
The haplotype method for discovering maternal grandsires (MGS) is an alternative to comparing one SNP at-a-time. For animals with an unknown dam or unlikely or unknown MGS and a confirmed sire, the haplotype method is employed as part of the weekly evaluation. The animal’s genotype is separated into the paternal and maternal contributions, then the maternal haplotypes are compared to the haplotypes of bulls that were born long enough before the animal to be a possible grandsire The true MGS is expected to have 45% of its haplotypes in common, allowing for the effect of recombination. A bull is proposed as the MGS if his percentage of matches is at least 15% higher than the bull with the next highest percentage of matches. If the pedigree MGS was designated as unlikely and haplotype discovery process identifies the pedigree MGS as the likely MGS, the unlikely designation is removed.
Timing: Haplotype based analysis is run as part of the weekly and monthly genomic evaluations. Genotypes with a confirmed sire are included in the weekly even if they do not qualify for evaluation.
Update program¶
CDCB runs an update program twice a day (5:00am and noon) to update the genomic information that was changed between the two runs.
For example, even though some pedigree information is updated to resolve a genomic conflict, the genomic conflict will not be cleared until the update program ends.
If a genotype status is not changed, even after the correction, the nominator should follow the checks below before contacting CDCB:
1) Check if the correction was accepted successfully (without any errors)
2) Check if the genotype is error-free
3) Wait for the next update program runs at 5:00am or noon
4) if all 3 above were checked but the genomic error has not been cleared or the genotype does not become usable, then contact CDCB customer service via Redmine ticket
USA or 840 for American country code?¶
A general rule is that IDs with ID number higher than 003001000002 should have a country code of 840, whereas ID numbers lower than 003001000002 should have an USA country code.
The country code of 840 was introduced with the implementation of RFID to avoid modifying the ID. Since there was some overlap in the range of numbers between 840 and USA IDs, this policy ensures uniqueness of IDs. It is important that 840/USA are accurately indicated in the animalID as those IDs are the standards shared nationally and internationally.
- Ex:
IDs starting with 840: HO840003001000003
IDs starting with USA: HOUSA003001000001
Zero Key¶
Zero Key is assigned automatically to a genotype for which a matching to a valid animal ID was not possible. This is usually caused by missing pedigree and/or nomination.
Please contact the CDCB genomics team if there is any other information you would like to add to this page.